While we’re currently being flooded by papers on the
intestinal microbiome, we still have very few dealing with the intestinal “micro-eukaryome”
(forgive me my "badomics", I should have known better after reading this piece by Dr Eisen).
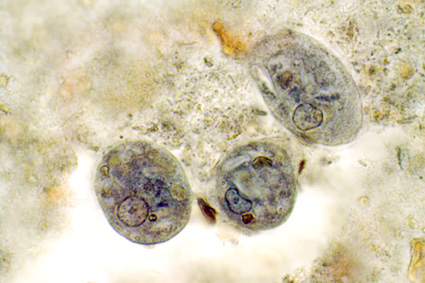
Hamad et al., just published their work on “Molecular Detection of Eukaryotes in a Single Human Stool Sample from Senegal” in PLoS One. They used a panel of 22 broad-specificity eukaryotic primers on genomic DNA extracted directly from faeces, cloned PCR products and did a blast search of the resulting sequences. They found about 18 micro-eukaryotic species in this particular faecal sample, most of which were fungi, and only two of which were “parasites”, namely Blastocystis sp. (subtype not given) and Entamoeba hartmanni, a so-called non-pathogenic amoebic species.They used both culture and culture-independent methods (PCR directly on genomic DNA from faeces) for the detection of intestinal fungi.
The study is interesting for a number of reasons:
1) It is one of the few papers out there on micro-eukaryotic diversity in faecal samples (other ones are listed in the reading list below), and we still know very little about micro-eukaryotes' potential interaction with the host and their ecological niche.
2) Many fungal species were detected by cloning of PCR products obtained by various primer pairs. It is possible that many of these are fungi stemming from the environment and diet, and not actually fungi colonizing the intestinal tract of this person; indeed the primers were able to pick up eukaryotic DNA such as that from tomatoes and common hop, stemming from the person’s diet. This is also one of the draw-backs of studies of fungi in stool samples: Even for mycologists it may prove difficult to determine which fungi are likely to be colonisers rather than fungi in transit due to environmental exposure, including diet. Analysis of consecutive samples from the same individual(s) (similar to the approach by Scanlan and Marchesi (2008)) will assist in identifying which fungi are stable and probable colonisiers. Similar to other studies, the investigators highlight the disparate findings resulting from the use of culture-dependent and culture-independent analyses; culture may be a way of identifying which ones of the many fungi detected by PCR that are actual colonisers.
3) We still don’t know much about what to expect when we take an approach like this. In the present study, multiple primer pairs were put into use, and 11 primer pairs yielded PCR products. The primer pairs amplified products of different lengths (some of them covering the complete SSU rDNA (18S)), and large products can sometimes be difficult to amplify and/or sequence for a variety of reasons; also preferential amplification may be a limiting factor. What would sometimes be useful is an in-silico analysis of the spectrum of organisms covered – at least theoretically - by each set of primers. In the papers I’ve seen so far aiming to display the eukaryotic diversity in human stool, Blastocystis has been a consistent finding, while Dientamoeba fragilis, which, at least in Denmark is almost as prevalent as Blastocystis (in some cohorts even more prevalent) and can be seen in co-infection, has not been reported so far. When you are presented with a list like the one presented by Hamad et al., you are inclined to believe that this list is exhaustive, but I think in-silico analysis data on such broad-specificity primers used for the detection of eukaryotic DNA would help us validate the use of these primers. Another approach to test the applicability of this methodology is to construct samples of DNA from known organisms in different ratios... and then test how the primers and cloning perform. What is also important is the very method of DNA extraction... obviously, our ability to detect DNA from any organism relies on our ability to extract DNA from it.
4) The study of micro-eukaryotes and their roles in health and disease includes first and foremost knowledge about which species and lineages that can be found and which ones that are the most common. Molecular methods are needed to identify the organisms in our intestine, since for instance parasites that look the same (morphological identity) can be genetically diverse with differing abilities to cause disease. We know from studies of micro-eukaryotes in ruminants that for instance some ciliates can be directly beneficial to the host, while others - such as cryptosporidia - are virtually obligate pathogens causing watery diarrhoea. Moreover, some organisms, including micro-eukaryotes, may be extremely difficult to culture even short-term, and also microscopy has limitations.
While we are still searching for virulence genes and other effector proteins in common micro-eukaryotes such as Blastocystis and Dientamoeba fragilis which could potentially cause disease directly, we also need to look for more indirect effects. Although much lower in numbers than our bacteria, (some) micro-eukaryotes may predate on beneficial bacteria to an extent where dysbiosis is reached. "Defaunation" of the intestine is speculated to be associated not only with impaired absorption of nutrients, but also with the development of severe disesases such as colon cancer and if micro-eukaryotes are able to skew our flora, this may have indirect impact on our health; many of our commensal bacteria are essential to some of our vital body functions, - indeed our intestinal flora can be viewed as a separate organ (see previous blog posts).
In the era of "omics" and "ngs" tools, it is interessesting to see a paper on global microbiotic diversity using a "conventional" cloning and sequencing approach in 2012. It may be one of the last papers of its kind?
To sum up: it is clear that a healthy intestine may be populated by a variety of micro-eukaryotes and future studies of the structure and function of the intestinal microbiome including micro-eukaryotes will help us understand their role in health and disease.
Let me end this post by uploading an image depicting "A Tree of Eukaryotes" (including Blastocystis) from an excellent protist blog by a colleague - my rendition here is practically useless, but I hope it might tease you to go and look at it in detail on "Welcome to the Ocelloid" by Psi Wavefunction.
Further reading:
Hamad I, Sokhna C, Raoult D, & Bittar F (2012). Molecular detection of eukaryotes in a single human stool sample from senegal. PloS one, 7 (7) PMID: 22808282
Pandey PK, Siddharth J, Verma P, Bavdekar A, Patole MS, & Shouche YS (2012). Molecular typing of fecal eukaryotic microbiota of human infants and their respective mothers. Journal of biosciences, 37 (2), 221-6 PMID: 22581327
Scanlan PD, & Marchesi JR (2008). Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. The ISME journal, 2 (12), 1183-93 PMID: 18670396
Hamad et al., just published their work on “Molecular Detection of Eukaryotes in a Single Human Stool Sample from Senegal” in PLoS One. They used a panel of 22 broad-specificity eukaryotic primers on genomic DNA extracted directly from faeces, cloned PCR products and did a blast search of the resulting sequences. They found about 18 micro-eukaryotic species in this particular faecal sample, most of which were fungi, and only two of which were “parasites”, namely Blastocystis sp. (subtype not given) and Entamoeba hartmanni, a so-called non-pathogenic amoebic species.They used both culture and culture-independent methods (PCR directly on genomic DNA from faeces) for the detection of intestinal fungi.
The study is interesting for a number of reasons:
1) It is one of the few papers out there on micro-eukaryotic diversity in faecal samples (other ones are listed in the reading list below), and we still know very little about micro-eukaryotes' potential interaction with the host and their ecological niche.
2) Many fungal species were detected by cloning of PCR products obtained by various primer pairs. It is possible that many of these are fungi stemming from the environment and diet, and not actually fungi colonizing the intestinal tract of this person; indeed the primers were able to pick up eukaryotic DNA such as that from tomatoes and common hop, stemming from the person’s diet. This is also one of the draw-backs of studies of fungi in stool samples: Even for mycologists it may prove difficult to determine which fungi are likely to be colonisers rather than fungi in transit due to environmental exposure, including diet. Analysis of consecutive samples from the same individual(s) (similar to the approach by Scanlan and Marchesi (2008)) will assist in identifying which fungi are stable and probable colonisiers. Similar to other studies, the investigators highlight the disparate findings resulting from the use of culture-dependent and culture-independent analyses; culture may be a way of identifying which ones of the many fungi detected by PCR that are actual colonisers.
3) We still don’t know much about what to expect when we take an approach like this. In the present study, multiple primer pairs were put into use, and 11 primer pairs yielded PCR products. The primer pairs amplified products of different lengths (some of them covering the complete SSU rDNA (18S)), and large products can sometimes be difficult to amplify and/or sequence for a variety of reasons; also preferential amplification may be a limiting factor. What would sometimes be useful is an in-silico analysis of the spectrum of organisms covered – at least theoretically - by each set of primers. In the papers I’ve seen so far aiming to display the eukaryotic diversity in human stool, Blastocystis has been a consistent finding, while Dientamoeba fragilis, which, at least in Denmark is almost as prevalent as Blastocystis (in some cohorts even more prevalent) and can be seen in co-infection, has not been reported so far. When you are presented with a list like the one presented by Hamad et al., you are inclined to believe that this list is exhaustive, but I think in-silico analysis data on such broad-specificity primers used for the detection of eukaryotic DNA would help us validate the use of these primers. Another approach to test the applicability of this methodology is to construct samples of DNA from known organisms in different ratios... and then test how the primers and cloning perform. What is also important is the very method of DNA extraction... obviously, our ability to detect DNA from any organism relies on our ability to extract DNA from it.
4) The study of micro-eukaryotes and their roles in health and disease includes first and foremost knowledge about which species and lineages that can be found and which ones that are the most common. Molecular methods are needed to identify the organisms in our intestine, since for instance parasites that look the same (morphological identity) can be genetically diverse with differing abilities to cause disease. We know from studies of micro-eukaryotes in ruminants that for instance some ciliates can be directly beneficial to the host, while others - such as cryptosporidia - are virtually obligate pathogens causing watery diarrhoea. Moreover, some organisms, including micro-eukaryotes, may be extremely difficult to culture even short-term, and also microscopy has limitations.
While we are still searching for virulence genes and other effector proteins in common micro-eukaryotes such as Blastocystis and Dientamoeba fragilis which could potentially cause disease directly, we also need to look for more indirect effects. Although much lower in numbers than our bacteria, (some) micro-eukaryotes may predate on beneficial bacteria to an extent where dysbiosis is reached. "Defaunation" of the intestine is speculated to be associated not only with impaired absorption of nutrients, but also with the development of severe disesases such as colon cancer and if micro-eukaryotes are able to skew our flora, this may have indirect impact on our health; many of our commensal bacteria are essential to some of our vital body functions, - indeed our intestinal flora can be viewed as a separate organ (see previous blog posts).
In the era of "omics" and "ngs" tools, it is interessesting to see a paper on global microbiotic diversity using a "conventional" cloning and sequencing approach in 2012. It may be one of the last papers of its kind?
To sum up: it is clear that a healthy intestine may be populated by a variety of micro-eukaryotes and future studies of the structure and function of the intestinal microbiome including micro-eukaryotes will help us understand their role in health and disease.
Let me end this post by uploading an image depicting "A Tree of Eukaryotes" (including Blastocystis) from an excellent protist blog by a colleague - my rendition here is practically useless, but I hope it might tease you to go and look at it in detail on "Welcome to the Ocelloid" by Psi Wavefunction.
Further reading:
Hamad I, Sokhna C, Raoult D, & Bittar F (2012). Molecular detection of eukaryotes in a single human stool sample from senegal. PloS one, 7 (7) PMID: 22808282
Pandey PK, Siddharth J, Verma P, Bavdekar A, Patole MS, & Shouche YS (2012). Molecular typing of fecal eukaryotic microbiota of human infants and their respective mothers. Journal of biosciences, 37 (2), 221-6 PMID: 22581327